There's been a lot of hooplah in the last year or so about the fact that our ability to generate sequence has scaled faster than Moore's Law over the last few years, and the attendant challenges of scaling analysis capacity; see Figure 1a and 1b, this reddit discussion, and also my the sky is falling! blog post.
There's also been been some backlash -- it's gotten to the point where showing any of the various graphs is greeted with derision, at least judging by the talk-associated Twitter feed.

Figure 1a. DNA sequencing costs, from http://www.genome.gov/sequencingcosts/
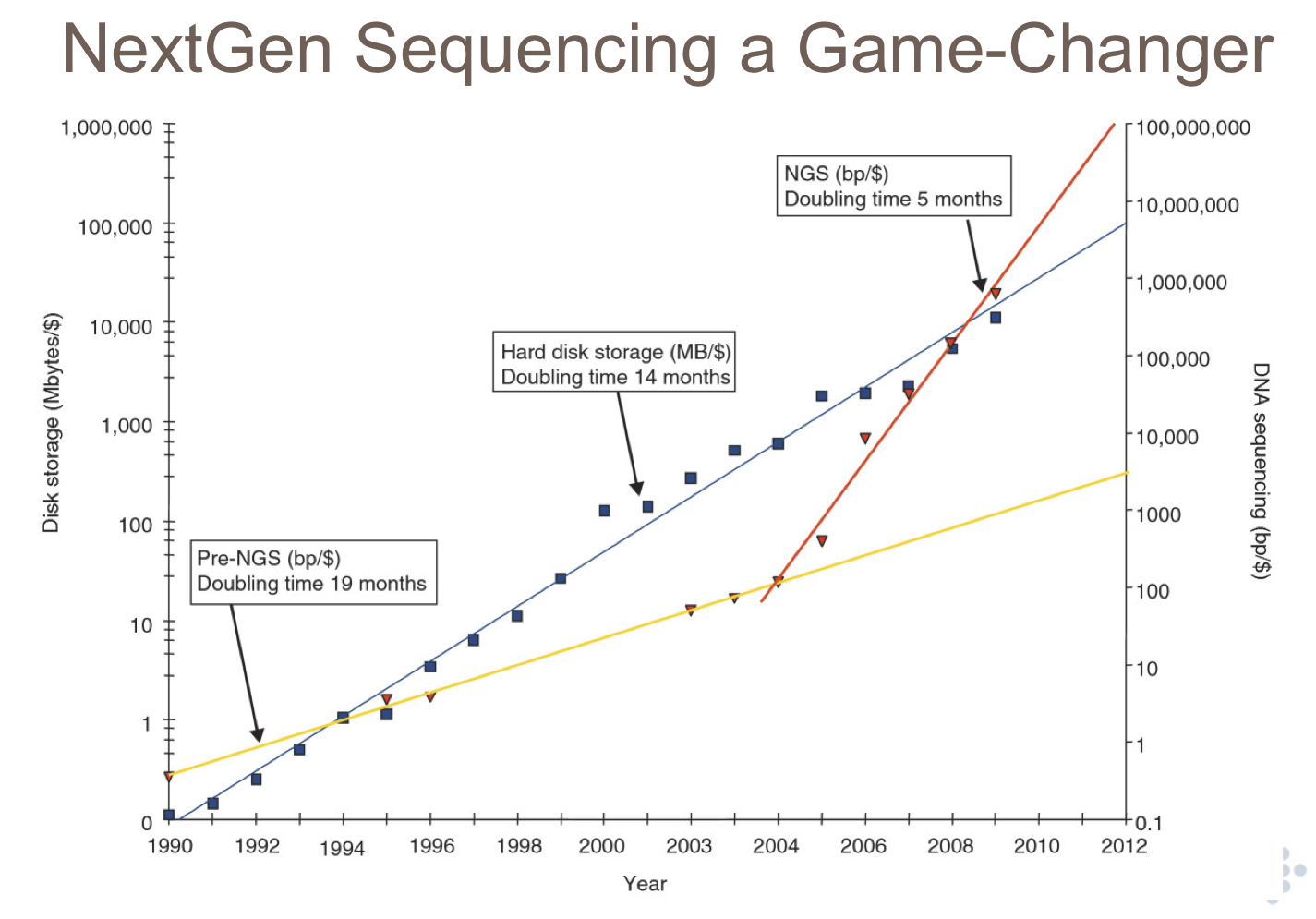
Figure 1b. Sequencing costs vs hard disk costs. Slide courtesy of Lincoln Stein.
From by the discussions I've seen, I think people still don't get -- or at least don't talk about -- the implications of this scaling behavior. In particular, I am surprised to hear the cloud (the cloud! the cloud!) touted as The Solution, since it's clearly not a solution to the actual scaling problem. (Also see: Wikipedia on cloud computing.)
To see why, consider the following model. Take two log-linear plots, one for cost per unit of compute power (CPU cycles, disk space, RAM, what have you), and one for cost per unit of sequence (\$\$ per bp of DNA). Now suppose that sequence cost is decreasing faster than compute cost, so you have two nice, diverging linear lines when you plot these trends over time on a log-linear plot (see Figure 2).

Figure 2. A simple model of the (exponential) decrease in compute costs vs (exponential) decrease sequencing data costs, against time.
Suppose we're interested in how much money to allocate to sequencing, vs how much money to allocate to compute -- the heart of the problem. How do these trends behave? One way to examine them is to look at the ratio of the data points.

Figure 3. The ratio of compute power to data over time, under the model in the previous figure.
As you'd expect, the ratio of compute power to data is also log-linear (Figure 3) -- it's just the difference between the two lines in Figure 2. Straight lines on log-linear plots, however, are in reality exponential -- see Figure 4! This is a linear-scale plot of compute costs relative to data costs -- and as you can see, compute costs end up dominating.

Figure 4. Ratio of compute power to data, over time, on a linear plot.
With this model, then, for the same dollar value of data, your relative compute costs will increase by a factor of 1000 over 10 years. This is true whether or not you're using the cloud! While your absolute costs may go up or down depending on infrastructure investments, Amazon's pricepoint, etc., the fundamental scaling behavior doesn't change. It doesn't much matter if Amazon is 2x cheaper than your HPC -- check out Figures 5a, b, and c if you need graphical confirmation of the math.



Figure 5a,b,c: Scaling behavior isn't affected by linearly lower costs.
The bottom line is this: when your data cost is decreasing faster than your hardware cost, the long-term solution cannot be to buy, rent, borrow, beg, or steal more hardware. The solution must lie in software and algorithms.
People who claim that cloud computing is going to provide an answer to the scaling issue with sequence, then, must be operating with some additional assumptions. Maybe they think the curves are shifted relative to one another, so that even 1000x costs are not a big deal - although figure 1 sort of argues against that. Like me, maybe they've heard that hard disks are about to start scaling way, way better -- if so, awesome! That might change the curves for data storage, if not analysis. Perhaps their research depends on using only a bounded amount of sequence -- e.g. single-genome sequencing, for which you can stop generating data at a certain point. Or perhaps they're proposing to use algorithms that scale sub-linearly with the amount of data they're applied to (although I don't know of any). Or perhaps they're planning for the shift in Moore's Law behavior that will come when that Amazon and other cloud computing providers build self-replicating compute clusters on the moon (hello, exo-atmospheric computing!) Whatever the plan, it would be interesting to hear their assumptions explained.
I think one likely answer to the Big Data conundrum in biology is that we'll come up with cleverer and cleverer approaches for quickly throwing away data that is unlikely to be of any use. Assuming these algorithms are linear in their application to data, but have smaller constants in front of their big-O, this will at least help stem the tide. (It will also, unfortunately, generate more and nastier biases in the results...) But I don't have any answers for what will happen in the medium term if sequencing continues to scale as it does.
It's also worth noting that de novo assembly (my current focus...) remains one of the biggest challenges. It requires gobs of the most expensive computational resource (RAM, which is not scaling as fast as disk and CPU), and there are no good solutions on the horizon for making it scale faster. Neither mRNAseq nor metagenomics are well-bounded problems (you always want more sequence!), and assembly will remain a critical approach for many people for many years. Moreover, cloud assembly approaches like Contrail are (sooner or later) doomed by the same logic as above. But it's a problem we need to solve! As I said at PyCon, "Life's too short to tackle the easy problems -- come to academia!".
--titus
p.s. If you want to play with the curves yourself, here's a Google Spreadsheet, and you can grab a straight CSV file here.
Legacy Comments
Posted by Deepak Singh on 2011-08-07 at 16:21.
Couldn't agree more. In some ways trying to forklift non-scalable applications and software to scalable infrastructure can be even more of a fools errand at scale.
Posted by Mitch Skinner on 2011-08-07 at 20:50.
I haven't heard people saying that the cloud is the One Solution that solves the problem forever, not even in the linked article by Stein. It's mostly about a few constant factors that make moving to the cloud potentially advantageous right now. In academia, if you're training people with skills that you hope will last them 40 years, then of course you have to ruthlessly exclude constant factors and focus only on asymptotic growth and longer-term trends. But if you're trying to get something done **this week**, then constant factors matter a lot.
Posted by Titus Brown on 2011-08-08 at 00:24.
Mitch, I'm not sure I agree that this is understood. We had a bunch of students read the Pennisi paper and most of them concluded "...the cloud is a solution". I'd say that at the least the magnitude of the challenge has not been discussed clearly, and in the absence of that explicit discussion I don't think non-CS folk get the point of how scaling works. NIH and NSF also aren't doing much (yet) to fund software development to the necessary degree, IMO, which makes me think that a lot of people don't really get it. I do address your point about constant factors at the end -- but I think we are well past that point of intersection; I can't assemble sequence data we got last year, much less what we're getting this year.
Posted by Mitch Skinner on 2011-08-08 at 02:08.
Fair enough. Given the data deluge, more funding for software development would be great, absolutely. I didn't realize you had quite so much data to assemble, which I suppose is your point. Re: assumptions (speaking only for me FWIW), I guess I've been mainly exposed to problems that can be usefully addressed with a Moore's-law level of scaling (plus, potentially, a cloud-supplied constant factor). So yes, I think there's plenty to do even with "only a bounded amount of sequence" (bounded above by Moore's law). But of course you're right that some problems are more difficult scaling- wise. I've also been assuming/hoping that new lab-side developments will help. For example, if Pac Bio or Oxford Nanopore or whoever comes out with decent long-read single-molecule sequencing, then some of the current hard problems change dramatically, right?
Posted by Titus Brown on 2011-08-08 at 11:03.
I see long reads as improving quality (esp of assembly!) but the data quantity will still be huge and present many of the same problems. I don't think mRNAseq or metagenomics of soil will change much with long reads because they still need really deep sampling. Hope I'm wrong ;)
Posted by Deepak Singh on 2011-08-08 at 13:38.
There are two parts to the cloud. One is elasticity and on-demand availability. That's the agility factor (and long term cost factor) that people flock to, and is important. The thing people forget is that scaling isn't magic. The cloud handles the operational challenges of scale,but software architecture still needs to cmoe from the community. The cloud architecture comes from a deep understanding of distributed systems, and until the life science community takes the route of rethinking software architecture, scale will always remain a challenge.
Posted by Rick Westerman on 2011-08-11 at 10:19.
In regards to the first two graphs, I think that they get derided because the data and/or assumptions in them is wrong. The first graph is particularly erroneous in that Moore's law (a simple exponential curve that is only about the number of transistors) is compared to costs -- it is an "apple to oranges" comparison. My own 10-years computing cost in our NGS sequencing center shows that computing costs do not follow Moore's law either. The NGS cost and computing cost curves do diverge but not as dramatically. In the second graph the assumption is hard drives reflect overall computing costs. However the above complaint should not take away from the need for better software. Almost all of the decreased cost in computing comes from parallel or clustered resources. If my bioinformatics program can not take advantage of such resources then, yes, indeed decreasing NGS costs will overwhelm my computing capabilities.
Posted by Titus Brown on 2011-08-11 at 12:01.
Rick, good comments! I pretty much agree with that :)
Comments !