At the NIH ADDS meeting, we had several breakout sessions. Michelle Dunn and I led the training session. For this breakout, we had the following agenda:
First, build "sticky-note clouds", with one sticky-note cloud with notes for each of the following topics:
- Desired outcomes of biomedical data science training;
- Other relevant training initiatives and connections;
- Topics that are part of data science;
- Axes of issues (e.g. online training, vs in person?)
Second, coalesce and summarize each topic;
Third, refine concepts and issues;
and fourth, come back with:
- three recommended actionable items that NIH can do _now_
- three things that NIH should leave to others (but be aware of)
This was not the greatest agenda, because we ended up spending only a very little time at the end talk about specific actionable items, but otherwise it was a great brainstorming session!
You can read the output document here; below, I expand and elaborate on what _I_ think everything meant.
Desired outcomes:
In no particular order,
- Increased appreciation by biomedical scientists of the role that data science could play;
- More biomedical scientists biologists using data science tools;
- More data scientists developing tools for biology;
- Increased cross-training of data scientists in biology;
- Map(s) and guidebooks to the data science landscape;
- Increased communication between biomedical scientists and data scientists;
- Making sure that all biomed grad students have a basic understanding of data science;
- Increased data science skills for university staff who provide services
Data science topics list
Data science topics list:
Stat & experimental design, modeling, math, prob, stochastic processing, NLP/text mining, Machine learning, inference, viz for exploration, regression, classification, Graphical models, data integration
CS - principles of metadata, metadata format algorithms, databases, optimization, complexity, programming, scripting, data management, data wrangling, distributed processing, cloud computing, software development, scripting, Workflows, tool usage
Presentation of results, computational thinking, statistical thinking
We arranged these topics on two diagrams, one set on the "data analysis workflow" chart and one set on the "practical to abstract skill" chart.
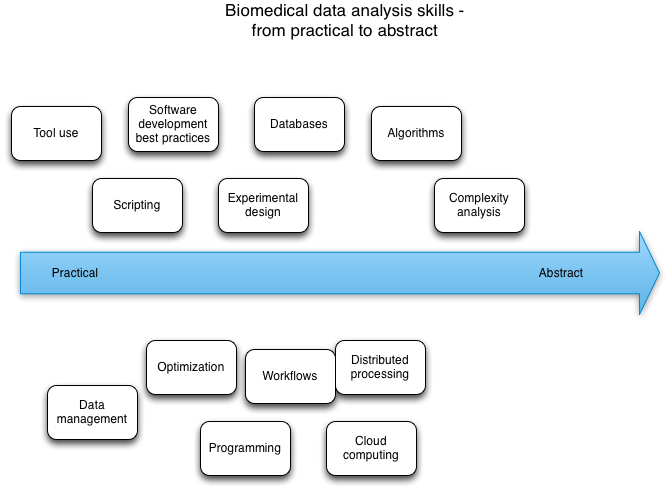
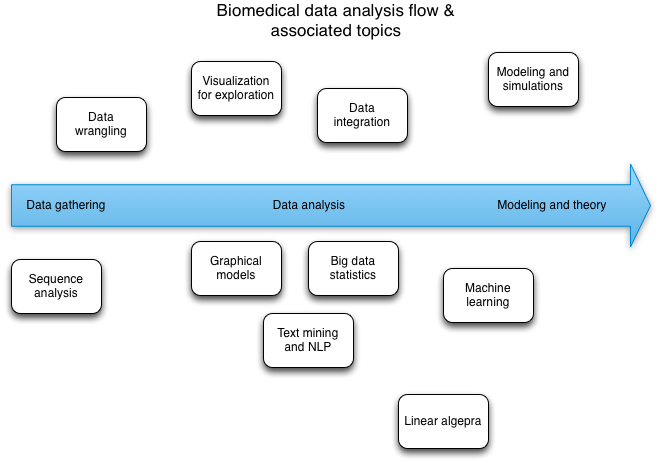
Patches welcome -- I did these in OmniGraffle, and you can grab the files here and here.
Axes/dichotomies of training:
The underlying question here was, how multidimensional is the biomedical training subspace? (Answer: D is around 10.)
- Clinical to biomedical to basic research - what topics do you target with your training?
- Just-in-time vs. background training - do you train for urgent research needs, or for broad background skills?
- Formal vs. informal - is the training formal and classroom oriented, or more informal and unstructured?
- Beginner vs. advanced - do you target beginner level or advanced level students?
- In-person vs. online - do you develop online courses or in-person courses?
- Short vs. long - short (two-day, two week) or long (semester, longer) or longer (graduate career) training?
- Centralized physically vs. federated/distributed physically - training centers, to which everyone travels? Or do you fly the trainers to the trainees? Or do you train the trainers and then have them train locally?
- Andragogy v. pedagogy (Full professor through undergrads) - do your target more senior or more junior people?
- Project-based vs. structured - do you assign open projects, or fairly stereotyped ones, or just go with pure didactic teaching?
- Individual learning vs team learning - self-explanatory.
I should say that I, at least, have no strong evidence that any one point on any of these spectra will be a good fit for even a large minority of potential trainees and goals; we're just trying to lay out the options here.
Existing training initiatives and data science connections:
- data science MOOCs, generally.
- Software Carpentry and Data Carpentry
- ELIXIR and especially ELIXIR-UK
- the NSF BIO Centers have several coordination grants that have converged on training (Data Carpentry emerged from two of them)
- Training NSF RCNs
- the Moore/Sloan Data Science Environments (UC Berkeley BIDS, , NYU CDS, University of Washington
- GOBLET
- The various QIIME Workshops that Rob Knight and Greg Caporaso run
- Existing and upcoming funded NIH R25s, including MBL STAMPS, and MSU NGS.
- More generally, any of these bioinformatics courses
Recommendations:
(The first two were from the group; the last three were from the meeting more broadly)
Solicit “crazier” R25s in existing BD2K framework; maybe open up another submission round before April.
Provide a list of topics and axes from our discussion; fund things like good software development practice, group-project camp for biomedical data driven discovery, development of good online data sets and explicit examples, hackathons, data management and metadata training, software development training, and communication training.
Solicit proposals for training coordination cores (2-3) to gather, catalog, and annotate existing resources, cooperate nationally and internationally, and collaborate on offering training. Also, develop evaluation and assessment criteria that can be applied across workshops.
CTB: fund early stage more broadly; don’t just limit training to graduate students in biomedical fields. For example, the biology grad student of today is the biomedical post doc of tomorrow.
Mercè Crosas: Fund internships, hands-on collaborative contributions in multi-disciplinary Data Science Labs
Leverage T-32 existing by adding req to include data science. (Addendum: there are already supplements available to do this)
Right now we're evaluating education grants individually; it would be good to also have a wide and deep evaluation done across grants.
A few other comments --
Training should cover senior and junior people
About 3% of the NIH budget is spent on training
Also, Daniel Mietchen (via online document comment) said: Consider using open formats. A good example is the Knight Challenge, currently asking for proposals around the future of libraries: https://www.newschallenge.org/challenge/libraries/brief.html
and that's all, folks!
--titus
Comments !